
Structures can be analyzed in a semi-automated way with scripting support. Measurement of bond distances and angles is straightforward in PyMOL. Obtain the phi and psi angles for your objects. Each command usually comes with one or more settings. which is called three-arm turnstile rotation. PyMOL can produce high-quality graphics, on par with Molscript, without needing to manually edit text files.
PYMOL TUTORIAL CHAIN FREE
It is fully extensible and available free to everyone via the “Python” license. Polyview-3D generates its images and animations at truly publication quality using PyMOL. The orange plane is part of the between Ile and Leu (blue plane), and the angle between the blue and yellow planes is phi. PIV displays the side-chain that is currently selected and the dihedral angles of the side-chain.

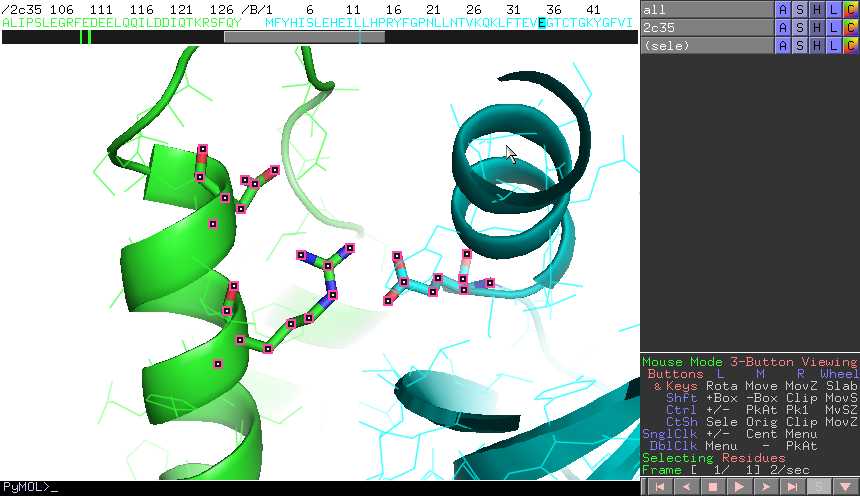
For this start PyMOL and in the command line window of PyMOL (indicated by PyMOL>) type: fetch 1F3G show cartoon hide lines show sticks, resn HIS. Then how … The second cluster represents the torsion angles between the fifth and sixth amino acids, and so on. 6-b and these values for M pro-LBD residues are listed in Table 1. Dihedral angles are defined for 4 atoms, so you are expected to choose the 4 atoms for measuring the angle. label_angle_digits sets the precision of angle labels Usage # set angle labels to display 2 decimals places to the right of the period set label_angle_digits, 2. Set the view to a wireframe rendering, by clicking 'S' next to 'all' in the display panel, hover over 'as', move to the other menu and click 'wire'. The conformation of the backbone can be described by two dihedral angles per residue, because the backbone residing between two juxtaposing Cα atoms are all in a single plane. How does proteins form different conformations and domains with the same 20 amino aicids. and the menu cascades to show the Measurement Mode choices. Try to locate the histidines in this structure. When you have completed the measurement the angle can be deleted by selecting Delete Last Object or left on the display screen. 8 7 4 3 0.0 S 72 5.0.The dihedral we want to parametrize is defined by these atoms (8-7-4-3), the starting angle is 0.0 and we want it to scan (S) for 72 steps with a step-size of 5 degrees (from 0 to 360). Given the structure above, provide a rough estimate of the value of the dihedral angle that describes the rotation of the central carbon-carbon bond in butane. Visual exploration of variable dihedral angles with Kinemage In my spare time I develop some free software. Cheers, Doug Quoting on Thu, 01:06:04 -0700: > Hello, PyMOL users! Natural linkers act as spacers between the domains of multidomain proteins. Interactive display and refinement of the predictions. "Take a set of different conformations and use principal component analysis on the phase space consisting of the dihedral angles." PyMOL is particularly attractive to us, since it has excellent features for viewing, it is fast and the display quality is superb, it can handle multiple molecules at once, and it is easy to define custom objects such as complexes or sets of atoms. I suggest writing a Python script that uses this function along with cmd.iterate() to loop over residues. By checking the Show Dots checkbox, contact dots are shown between the current side-chain and the rest of the protein. Pymol You should see a backbone representation of the protein with only the histidine side-chains visible. View Pymol Introduction Lab Worksheet W21.pdf from BIOG 2017 at Cornell University. Answer (1 of 2): Despite the fact that the alpha chain is on 16 and the beta chain is on 11 in humans, the alpha and beta chains are closely homologous and share the same fold.
PYMOL TUTORIAL CHAIN DOWNLOAD
This model, "butane.pse", is available for download on Canvas from the PyMOL page under Homework 8 Files. The PyMOL Layout! set ray_shadow, off #影を消す Pymol For example, the ray command comes with 50 settings! set_dihedral atom1, atom2, atom3, atom4, angle PYMOL API. The dihedral angles along a polypeptide chain are of three types: PyMol "dihedral" command generates a label to show the dihedral angle formed by 4 picked atoms.


 0 kommentar(er)
0 kommentar(er)
